SF_DECONVOLVE Documentation¶
This code implements the algorithm described in Farrens et al. (2017) to deconvolve images using sparsity or low-rank approximation regularisation. A short summary of this paper is available here.
Contents¶
Introduction¶
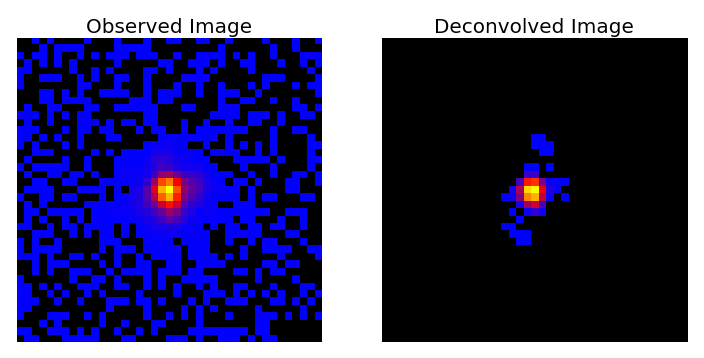
This repository contains a Python code designed for PSF deconvolution and analysis.
The directory lib contains several primary functions and classes, but the
majority of the optimisation and analysis tools are provided in
sf-tools.
Package Contents¶
Dependencies¶
Required Packages¶
In order to run the code in this repository the following packages must be installed:
- Python [Tested with v 2.7.11 and 3.6.3]
- Numpy [Tested with v 1.13.3]
- Scipy [Tested with v 0.18.1]
- Future [Tested with v 0.16.0]
- Astropy [Tested with v 1.3]
- sf-tools [Tested with v 1.0]
- The current implementation of wavelet transformations additionally requires
the
mr_transform.ccC++ script from the Sparse2D library in the iSAP package [Tested with v 3.1]. These C++ scripts will be need to be compiled in order to run (see iSAP Documentation for details). Note: The low-rank approximation method can be run purely in Python without the Sparse2D binaries.
Optional Packages¶
The following packages can optionally be installed to add extra functionality:
- Matplotlib [Tested with v 2.0.2]
- Termcolor [Tested with v 1.1.0]
Execution¶
The primary code is an executable script called sf_deconvolve.py which is
designed to take an observed (i.e. with PSF effects and noise) stack of
galaxy images and a known PSF, and attempt to reconstruct the original images.
The input format are Numpy binary files (.npy) or FITS image files (.fits).
Input Format¶
The input files should have the following format:
- Input Images: This should be either a Numpy binary or a FITS file containing a 3D array of galaxy images. e.g. for a sample of 10 images, each with size 41x41, the shape of the array should be [10, 41, 41].
- Input PSF(s): This should be either a Numpy binary or a FITS file containing a 2D array (for a fixed PSF) or a 3D array (for a spatially varying PSF) of PSF images. For the spatially varying case the number of PSF images must match the number of corresponding galaxy images. e.g. For a sample of 10 images the codes expects 10 PSFs.
See the files provided in the examples directory for reference.
Running the executable script¶
The code can be run in a terminal (not in a Python session) as follows:
$ sf_deconvolve.py -i INPUT_IMAGES.npy -p PSF.npy -o OUTPUT_NAME
Where INPUT_IMAGES.npy denotes the Numpy binary file containing the stack
of observed galaxy images, PSF.npy denotes the PSF corresponding to each
galaxy image and OUTPUT_NAME specifies the output path and file name.
Alternatively the code arguments can be stored in a configuration file (with
any name) and the code can be run by providing
the file name preceded by a @.
$ sf_deconvolve.py @config.ini
An example configuration file is provided in the examples directory.
Running the code in a Python session¶
The code can be run in an active Python session in two ways. For either
approach first import sf_deconvolve:
>>> import sf_deconvolve
The first approach simply runs the full script where the command line arguments can be passed as a list of strings:
>>> sf_deconvolve.main(['-i', 'INPUT_IMAGES.npy', '-p', 'PSF.npy', '-o', 'OUTPUT_NAME'])
The second approach assumes that the user has already has read the images and PSF(s) into memory and wishes to return the deconvolution results to memory:
>>> opts = vars(sf_deconvolve.get_opts(['-i', 'INPUT_IMAGES.npy', '-p', 'PSF.npy', '-o', 'OUTPUT_NAME']))
>>> primal_res, dual_res, psf_res = sf_deconvolve.run(INPUT_IMAGES, INPUT_PSFS, **opts)
Where INPUT_IMAGES and INPUT_PSFS are both Numpy arrays. The resulting
deconvolved images will be saved to the variable primal_res.
In both cases it is possible to read a predefined configuration file.
>>> opts = vars(sf_deconvolve.get_opts(['@config.ini']))
>>> primal_res, dual_res, psf_res = sf_deconvolve.run(INPUT_IMAGES, INPUT_PSFS, **opts)
Example¶
The following example can be run on the sample data provided in the example
directory.
This example takes a sample of 100 galaxy images (with PSF effects and added noise) and the corresponding PSFs, and recovers the original images using low-rank approximation via Condat-Vu optimisation.
$ sf_deconvolve.py -i example_image_stack.npy -p example_psfs.npy -o example_output --mode lowr
The example can also be run using the configuration file provided.
The result will be two Numpy binary files called example_output_primal.npy
and example_output_dual.npy corresponding to the primal and dual variables
in the splitting algorithm. The reconstructed images will be in the
example_output_primal.npy file.
The example can also be run with the FITS files provided.
Code Options¶
Required Arguments¶
- -i INPUT, –input INPUT: Input data file name. File should be a Numpy binary containing a stack of noisy galaxy images with PSF effects (i.e. a 3D array).
- -p PSF, –psf PSF: PSF file name. File should be a Numpy binary containing either: (a) a single PSF (i.e. a 2D array for fixed format) or (b) a stack of PSFs corresponding to each of the galaxy images (i.e. a 3D array for obj_var format).
Optional Arguments¶
- -h, –help: Show the help message and exit.
- -v, –version: Show the program’s version number and exit.
- -q, –quiet: Suppress verbose for each iteration.
- -o, –output: Output file name. If not specified output files will placed in input file path.
- –output_format Output file format [npy or fits].
Initialisation:
- -k, –current_res: Current deconvolution results file name (i.e. the file containing the primal results from a previous run).
- –noise_est: Initial estimate of the noise standard deviation in the observed galaxy images. If not specified this quantity is automatically calculated using the median absolute deviation of the input image(s).
Optimisation:
- -m, –mode {all,sparse,lowr,grad}: Option to specify the optimisation mode [all, sparse, lowr or grad]. all performs optimisation using both low-rank approximation and sparsity, sparse using only sparsity, lowr uses only low-rank and grad uses only gradient descent. (default: lowr)
- –opt_type {condat,fwbw,gfwbw}: Option to specify the optimisation method to be implemented [condat, fwbw or gfwbw]. condat implements the Condat-Vu proximal splitting method, fwbw implements Forward-Backward splitting with FISTA speed-up and gfwbw implements the generalised Forward-Backward splitting method. (default: condat)
- –n_iter: Number of iterations. (default: 150)
- –cost_window: Window to measure cost function (i.e. interval of iterations for which cost should be calculated). (default: 1)
- –convergence: Convergence tolerance. (default: 0.0001)
- –no_pos: Option to turn off positivity constraint.
- –grad_type: Option to specify the type of gradient [psf_known, psf_unknown, none]. psf_known implements deconvolution with PSFs provided, psf_unknown simultaneously improves the PSF while performing the deconvolution, none implements deconvolution without gradient descent (for testing purposes only). (default: psf_known)
Low-Rank Aproximation:
- –lowr_thresh_factor: Low rank threshold factor. (default: 1)
- –lowr_type: Type of low-rank regularisation [standard or ngole]. (default: standard)
- –lowr_thresh_type: Low rank threshold type [soft or hard]. (default: hard)
Sparsity:
- –wavelet_type: Type of Wavelet to be used (see iSAP Documentation). (default: 1)
- –wave_thresh_factor: Wavelet threshold factor. (default: [3.0, 3.0, 4.0])
- –n_reweights: Number of reweightings. (default: 1)
PSF Estimation
- –lambda_psf: Regularisation control parameter for PSF estimation. (default: 1.0)
- –beta_psf: Gradient step for PSF estimation. (default: 1.0)
Condat Algorithm:
- –relax: Relaxation parameter (rho_n in Condat-Vu method). (default: 0.8)
- –condat_sigma: Condat proximal dual parameter. If the option is provided without any value, an appropriate value is calculated automatically. (default: 0.5)
- –condat_tau: Condat proximal primal parameter. If the option is provided without any value, an appropriate value is calculated automatically. (default: 0.5)
Testing:
- -c, –clean_data: Clean data file name.
- -r, –random_seed: Random seed. Use this option if the input data is a randomly selected subset (with known seed) of the full sample of clean data.
- –true_psf: True PSFs file name.
- –kernel: Standard deviation of pixels for Gaussian kernel. This option will multiply the deconvolution results by a Gaussian kernel.
- –metric: Metric to average errors [median or mean]. (default: median)
Troubleshooting¶
- If you get the following error:
ERROR: svd() got an unexpected keyword argument 'lapack_driver'
Update your Numpy and Scipy installations
$ pip install --upgrade numpy
$ pip install --upgrade scipy